Lecture 11 (4/20/2022)¶
Announcements
Problem set 3 coming out today, will be due next Wednesday 4/27
Last time we covered:
Data cleaning with python (duplicates, missing data, outliers)
Today’s agenda:
Wide versus long data
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
Wide and Long Data¶
“Like families, tidy datasets are all alike but every messy dataset is messy in its own way.” ~ Hadley Wickham
What is wide data?¶
…
When we interact with data that’s made to be read by people, it’s most often in wide format.
The definition of wide data can be a little hard to pin down but one rule of thumb is that wide data spreads multiple observations or variables across columns in a given row.
y |
x1 |
x2 |
x3 |
|---|---|---|---|
1 |
a |
b |
c |
2 |
d |
e |
f |
… |
… |
… |
… |
Here’s some data I made up about average temperatures in five US cities over three consecutive years:
cities = pd.DataFrame({
"City": ["San Diego", "Denver", "New York City", "Los Angeles", "San Francisco"],
"2010": [75, 60, 55, 65, 70],
"2011": [77, 63, 58, 67, 72],
"2012": [77, 62, 56, 67, 71]
})
cities
| City | 2010 | 2011 | 2012 | |
|---|---|---|---|---|
| 0 | San Diego | 75 | 77 | 77 |
| 1 | Denver | 60 | 63 | 62 |
| 2 | New York City | 55 | 58 | 56 |
| 3 | Los Angeles | 65 | 67 | 67 |
| 4 | San Francisco | 70 | 72 | 71 |
This data can also be presented with year as our variable of interest and each city as a column:
years = pd.DataFrame({
"Year": [2010, 2011, 2012],
"San Diego": [75, 77, 77],
"Denver": [60, 63, 62],
"New York City": [55, 58, 56],
"Los Angeles": [65, 67, 67],
"San Francisco": [70, 72, 71]
})
years
| Year | San Diego | Denver | New York City | Los Angeles | San Francisco | |
|---|---|---|---|---|---|---|
| 0 | 2010 | 75 | 60 | 55 | 65 | 70 |
| 1 | 2011 | 77 | 63 | 58 | 67 | 72 |
| 2 | 2012 | 77 | 62 | 56 | 67 | 71 |
Both of these are pretty easy to read and pretty intuitive.
What kind of questions can we answer most easily with each dataframe?
cities:
years:
Note: this is easiest to illustrate with time sequence data, but most data can be toggled around this way to some degree:
students = pd.DataFrame({
"Student": ["Erik", "Amanda", "Maia"],
"Math": [90, 95, 80],
"Writing": [90, 85, 95]
})
students
| Student | Math | Writing | |
|---|---|---|---|
| 0 | Erik | 90 | 90 |
| 1 | Amanda | 95 | 85 |
| 2 | Maia | 80 | 95 |
classes = pd.DataFrame({
"Subject": ["Math", "Writing"],
"Erik": [80, 95],
"Amanda": [95, 85],
"Maia": [80, 95]
})
classes
| Subject | Erik | Amanda | Maia | |
|---|---|---|---|---|
| 0 | Math | 80 | 95 | 80 |
| 1 | Writing | 95 | 85 | 95 |
The first table makes it easier to ask questions like “which student performed best?”, while the second is easier for asking questions like “are these students better at math or writing?”
Self-quiz: do the above examples give you an okay intuition for what wide data looks like?
So what’s the problem?¶
First, the exercise above suggests that for different kinds of questions, we need to format our data in different ways. That seems onerous…
Second, even though tables like this make these data easy to read as humans, answering questions about the data when it’s formatted like this can be difficult and inefficient.
Using the data below, how do we figure out which city was hottest on average (using our python skills)?
cities
| City | 2010 | 2011 | 2012 | |
|---|---|---|---|---|
| 0 | San Diego | 75 | 77 | 77 |
| 1 | Denver | 60 | 63 | 62 |
| 2 | New York City | 55 | 58 | 56 |
| 3 | Los Angeles | 65 | 67 | 67 |
| 4 | San Francisco | 70 | 72 | 71 |
# CODE HERE
Notice that we have to do this by calculating an average row by row. Seems inefficient.
Can we do any better with our years dataframe?
years
| Year | San Diego | Denver | New York City | Los Angeles | San Francisco | |
|---|---|---|---|---|---|---|
| 0 | 2010 | 75 | 60 | 55 | 65 | 70 |
| 1 | 2011 | 77 | 63 | 58 | 67 | 72 |
| 2 | 2012 | 77 | 62 | 56 | 67 | 71 |
# CODE HERE
Using the data below, how do we decide which year had the highest recorded temperature across these cities?
years
| Year | San Diego | Denver | New York City | Los Angeles | San Francisco | |
|---|---|---|---|---|---|---|
| 0 | 2010 | 75 | 60 | 55 | 65 | 70 |
| 1 | 2011 | 77 | 63 | 58 | 67 | 72 |
| 2 | 2012 | 77 | 62 | 56 | 67 | 71 |
# CODE HERE
Yikes 😬
Self-quiz: is it clear how data that’s easy to read in wide format can be kind of tricky to interact with when trying to analyze it in python?
What do we do about this? Answer: tidy (long) data!¶
With long or tidy data, every observation gets its own row, with columns indicating the variable values that correpond to that observation.
The wide table at the beginning of the previous section looked like this:
y |
x1 |
x2 |
x3 |
|---|---|---|---|
1 |
a |
b |
c |
2 |
d |
e |
f |
… |
… |
… |
… |
Compare the table above to this one:
y |
variable |
value |
|---|---|---|
1 |
x1 |
a |
1 |
x2 |
b |
1 |
x3 |
c |
2 |
x1 |
d |
2 |
x2 |
e |
2 |
x3 |
f |
… |
… |
… |
Here’s a concrete example with the student data above.
In wide form, it looked like this:
students
| Student | Math | Writing | |
|---|---|---|---|
| 0 | Erik | 90 | 90 |
| 1 | Amanda | 95 | 85 |
| 2 | Maia | 80 | 95 |
In tidy form, it looks like this:
tidy_students = pd.DataFrame({
"Student": ["Erik", "Erik", "Amanda", "Amanda", "Maia", "Maia"],
"Subject": ["Math", "Writing", "Math", "Writing", "Math", "Writing"],
"Score": [90, 90, 95, 85, 80, 95]
})
tidy_students
| Student | Subject | Score | |
|---|---|---|---|
| 0 | Erik | Math | 90 |
| 1 | Erik | Writing | 90 |
| 2 | Amanda | Math | 95 |
| 3 | Amanda | Writing | 85 |
| 4 | Maia | Math | 80 |
| 5 | Maia | Writing | 95 |
Self-quiz: is it clear how the tidy data here differs from wide data?
If you want to go into the weeds on this, here’s a paper by the inventor of tidyverse, a large library in R with many similar functions to pandas.
So what does tidy data do for us?¶
The tidy data in the previous examples are harder to read and harder to interpret in the ways we often want to think about tabular data.
So how does this help us?
Summary
Tidy data avoids the pitfalls of having to reformat our data for different kinds of questions (usually)
Tidy data enforces structure so there isn’t confusion about how best to represent our data (there may be multiple wide formats but usually only one tidy format) -> best practice
Tidy data is easier to interact with and analyze with code
Tidy data lets us take advantage of the vectorization that numpy, pandas, and other modern coding languages employ to make calculations super speedy
Example
Let’s go through a simple example with the temperature data above.
Here’s the original wide dataframe:
cities
| City | 2010 | 2011 | 2012 | |
|---|---|---|---|---|
| 0 | San Diego | 75 | 77 | 77 |
| 1 | Denver | 60 | 63 | 62 |
| 2 | New York City | 55 | 58 | 56 |
| 3 | Los Angeles | 65 | 67 | 67 |
| 4 | San Francisco | 70 | 72 | 71 |
Here it is in tidy format:
tidy_cities = pd.DataFrame({
"City": ["San Diego", "San Diego", "San Diego",
"Denver", "Denver", "Denver",
"New York City", "New York City", "New York City",
"Los Angeles", "Los Angeles", "Los Angeles",
"San Francisco", "San Francisco", "San Francisco"
],
"Year": [2010, 2011, 2012,
2010, 2011, 2012,
2010, 2011, 2012,
2010, 2011, 2012,
2010, 2011, 2012
],
"Temp": [75, 77, 77,
60, 63, 62,
55, 58, 56,
65, 67, 67,
70, 72, 71
]
})
tidy_cities
| City | Year | Temp | |
|---|---|---|---|
| 0 | San Diego | 2010 | 75 |
| 1 | San Diego | 2011 | 77 |
| 2 | San Diego | 2012 | 77 |
| 3 | Denver | 2010 | 60 |
| 4 | Denver | 2011 | 63 |
| 5 | Denver | 2012 | 62 |
| 6 | New York City | 2010 | 55 |
| 7 | New York City | 2011 | 58 |
| 8 | New York City | 2012 | 56 |
| 9 | Los Angeles | 2010 | 65 |
| 10 | Los Angeles | 2011 | 67 |
| 11 | Los Angeles | 2012 | 67 |
| 12 | San Francisco | 2010 | 70 |
| 13 | San Francisco | 2011 | 72 |
| 14 | San Francisco | 2012 | 71 |
Now, let’s return to our original question: which city was the hottest on average during this time?
# CODE HERE
That was pretty easy.
And under the hood, pandas groupby means that we compute the average temperature using vectorization rather than calculating row by row as we did in the solution above.
What about our second question: which year had the highest recorded temperature?
# CODE HERE
Okay, that was also pretty easy.
So, this is far from an exhaustive survey of wide versus tidy/long data, but should give you a flavor for why this distinction is useful.
Self-quiz: do the examples above make it pretty clear why tidy data makes our lives simpler, clearer, and easier for coding / analysis?
Pandas helps you convert data easily¶
Lots of data in the real world comes in wide form or requires some re-shuffling to get into tidy format.
If you’re working with a dataset that isn’t in tidy form, it’s almost always a good first step.
We’ll quickly review the tools that pandas has for toggling data formats.
Much more info about this here.
Converting from wide to long with melt¶
First, let’s turn to a familiar dataset: the gapminder data.
Is this data in tidy form?
gap = pd.read_csv("https://raw.githubusercontent.com/UCSD-CSS-002/ucsd-css-002.github.io/master/datasets/gapminder.csv")
gap
# gap.shape # note the size. Things are about to change...
| Unnamed: 0 | country | continent | year | lifeExp | pop | gdpPercap | |
|---|---|---|---|---|---|---|---|
| 0 | 1 | Afghanistan | Asia | 1952 | 28.801 | 8425333 | 779.445314 |
| 1 | 2 | Afghanistan | Asia | 1957 | 30.332 | 9240934 | 820.853030 |
| 2 | 3 | Afghanistan | Asia | 1962 | 31.997 | 10267083 | 853.100710 |
| 3 | 4 | Afghanistan | Asia | 1967 | 34.020 | 11537966 | 836.197138 |
| 4 | 5 | Afghanistan | Asia | 1972 | 36.088 | 13079460 | 739.981106 |
| ... | ... | ... | ... | ... | ... | ... | ... |
| 1699 | 1700 | Zimbabwe | Africa | 1987 | 62.351 | 9216418 | 706.157306 |
| 1700 | 1701 | Zimbabwe | Africa | 1992 | 60.377 | 10704340 | 693.420786 |
| 1701 | 1702 | Zimbabwe | Africa | 1997 | 46.809 | 11404948 | 792.449960 |
| 1702 | 1703 | Zimbabwe | Africa | 2002 | 39.989 | 11926563 | 672.038623 |
| 1703 | 1704 | Zimbabwe | Africa | 2007 | 43.487 | 12311143 | 469.709298 |
1704 rows × 7 columns
Let’s move the “observations” (lifeExp, pop, and gdpPercap) to their own rows using melt:
gap_tidy = gap.melt(
id_vars = ["Unnamed: 0", "country", "continent", "year"], # columns to keep in each row
value_vars = ["lifeExp", "pop", "gdpPercap"], # columns to be moved into their own rows
var_name = "measure", # name of the column that will store the "value_vars" column names
value_name = "value" # name of the column that will store the "value_vars" column values
)
gap_tidy # take a look at the data. Is this what you expected?
# gap_tidy.shape # note how many rows we added with this
| Unnamed: 0 | country | continent | year | measure | value | |
|---|---|---|---|---|---|---|
| 0 | 1 | Afghanistan | Asia | 1952 | lifeExp | 28.801000 |
| 1 | 2 | Afghanistan | Asia | 1957 | lifeExp | 30.332000 |
| 2 | 3 | Afghanistan | Asia | 1962 | lifeExp | 31.997000 |
| 3 | 4 | Afghanistan | Asia | 1967 | lifeExp | 34.020000 |
| 4 | 5 | Afghanistan | Asia | 1972 | lifeExp | 36.088000 |
| ... | ... | ... | ... | ... | ... | ... |
| 5107 | 1700 | Zimbabwe | Africa | 1987 | gdpPercap | 706.157306 |
| 5108 | 1701 | Zimbabwe | Africa | 1992 | gdpPercap | 693.420786 |
| 5109 | 1702 | Zimbabwe | Africa | 1997 | gdpPercap | 792.449960 |
| 5110 | 1703 | Zimbabwe | Africa | 2002 | gdpPercap | 672.038623 |
| 5111 | 1704 | Zimbabwe | Africa | 2007 | gdpPercap | 469.709298 |
5112 rows × 6 columns
What can we do with this?
Quick example!
(think about how we would do the below with our data in wide format)
gap_tidy.groupby(
['country', 'measure']
)['value'].mean().reset_index()
| country | measure | value | |
|---|---|---|---|
| 0 | Afghanistan | gdpPercap | 8.026746e+02 |
| 1 | Afghanistan | lifeExp | 3.747883e+01 |
| 2 | Afghanistan | pop | 1.582372e+07 |
| 3 | Albania | gdpPercap | 3.255367e+03 |
| 4 | Albania | lifeExp | 6.843292e+01 |
| ... | ... | ... | ... |
| 421 | Zambia | lifeExp | 4.599633e+01 |
| 422 | Zambia | pop | 6.353805e+06 |
| 423 | Zimbabwe | gdpPercap | 6.358580e+02 |
| 424 | Zimbabwe | lifeExp | 5.266317e+01 |
| 425 | Zimbabwe | pop | 7.641966e+06 |
426 rows × 3 columns
Converting from long to wide with pivot¶
But wait! I thought we wanted our data in tidy format???
The pivot function makes it easy for us to convert to wide format when it’s convenient.
gap_wide = gap_tidy.pivot(
index = "year", # column to be treated as the index
columns = ["measure", "country"], # columns to be spread out into their own columns for each value
values = "value" # value to be inserted in each new column
)
gap_wide
# gap_wide.shape # this is super condensed
| measure | lifeExp | ... | gdpPercap | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| country | Afghanistan | Albania | Algeria | Angola | Argentina | Australia | Austria | Bahrain | Bangladesh | Belgium | ... | Uganda | United Kingdom | United States | Uruguay | Venezuela | Vietnam | West Bank and Gaza | Yemen, Rep. | Zambia | Zimbabwe |
| year | |||||||||||||||||||||
| 1952 | 28.801 | 55.230 | 43.077 | 30.015 | 62.485 | 69.120 | 66.800 | 50.939 | 37.484 | 68.000 | ... | 734.753484 | 9979.508487 | 13990.48208 | 5716.766744 | 7689.799761 | 605.066492 | 1515.592329 | 781.717576 | 1147.388831 | 406.884115 |
| 1957 | 30.332 | 59.280 | 45.685 | 31.999 | 64.399 | 70.330 | 67.480 | 53.832 | 39.348 | 69.240 | ... | 774.371069 | 11283.177950 | 14847.12712 | 6150.772969 | 9802.466526 | 676.285448 | 1827.067742 | 804.830455 | 1311.956766 | 518.764268 |
| 1962 | 31.997 | 64.820 | 48.303 | 34.000 | 65.142 | 70.930 | 69.540 | 56.923 | 41.216 | 70.250 | ... | 767.271740 | 12477.177070 | 16173.14586 | 5603.357717 | 8422.974165 | 772.049160 | 2198.956312 | 825.623201 | 1452.725766 | 527.272182 |
| 1967 | 34.020 | 66.220 | 51.407 | 35.985 | 65.634 | 71.100 | 70.140 | 59.923 | 43.453 | 70.940 | ... | 908.918522 | 14142.850890 | 19530.36557 | 5444.619620 | 9541.474188 | 637.123289 | 2649.715007 | 862.442146 | 1777.077318 | 569.795071 |
| 1972 | 36.088 | 67.690 | 54.518 | 37.928 | 67.065 | 71.930 | 70.630 | 63.300 | 45.252 | 71.440 | ... | 950.735869 | 15895.116410 | 21806.03594 | 5703.408898 | 10505.259660 | 699.501644 | 3133.409277 | 1265.047031 | 1773.498265 | 799.362176 |
| 1977 | 38.438 | 68.930 | 58.014 | 39.483 | 68.481 | 73.490 | 72.170 | 65.593 | 46.923 | 72.800 | ... | 843.733137 | 17428.748460 | 24072.63213 | 6504.339663 | 13143.950950 | 713.537120 | 3682.831494 | 1829.765177 | 1588.688299 | 685.587682 |
| 1982 | 39.854 | 70.420 | 61.368 | 39.942 | 69.942 | 74.740 | 73.180 | 69.052 | 50.009 | 73.930 | ... | 682.266227 | 18232.424520 | 25009.55914 | 6920.223051 | 11152.410110 | 707.235786 | 4336.032082 | 1977.557010 | 1408.678565 | 788.855041 |
| 1987 | 40.822 | 72.000 | 65.799 | 39.906 | 70.774 | 76.320 | 74.940 | 70.750 | 52.819 | 75.350 | ... | 617.724406 | 21664.787670 | 29884.35041 | 7452.398969 | 9883.584648 | 820.799445 | 5107.197384 | 1971.741538 | 1213.315116 | 706.157306 |
| 1992 | 41.674 | 71.581 | 67.744 | 40.647 | 71.868 | 77.560 | 76.040 | 72.601 | 56.018 | 76.460 | ... | 644.170797 | 22705.092540 | 32003.93224 | 8137.004775 | 10733.926310 | 989.023149 | 6017.654756 | 1879.496673 | 1210.884633 | 693.420786 |
| 1997 | 41.763 | 72.950 | 69.152 | 40.963 | 73.275 | 78.830 | 77.510 | 73.925 | 59.412 | 77.530 | ... | 816.559081 | 26074.531360 | 35767.43303 | 9230.240708 | 10165.495180 | 1385.896769 | 7110.667619 | 2117.484526 | 1071.353818 | 792.449960 |
| 2002 | 42.129 | 75.651 | 70.994 | 41.003 | 74.340 | 80.370 | 78.980 | 74.795 | 62.013 | 78.320 | ... | 927.721002 | 29478.999190 | 39097.09955 | 7727.002004 | 8605.047831 | 1764.456677 | 4515.487575 | 2234.820827 | 1071.613938 | 672.038623 |
| 2007 | 43.828 | 76.423 | 72.301 | 42.731 | 75.320 | 81.235 | 79.829 | 75.635 | 64.062 | 79.441 | ... | 1056.380121 | 33203.261280 | 42951.65309 | 10611.462990 | 11415.805690 | 2441.576404 | 3025.349798 | 2280.769906 | 1271.211593 | 469.709298 |
12 rows × 426 columns
# We access data in the above by making our way down the hierarchical columns
gap_wide.columns
gap_wide['lifeExp']['Australia'][2002]
80.37
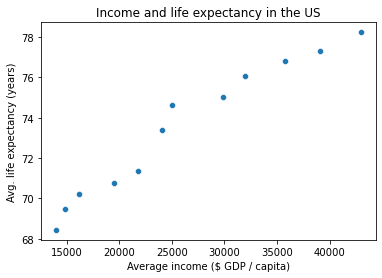
# This can make things like plotting this data a little easier (no need to filter ahead of time)
g = sns.scatterplot(x = gap_wide['gdpPercap']['United States'],
y = gap_wide['lifeExp']['United States']
)
g.set_xlabel("Average income ($ GDP / capita)")
g.set_ylabel("Avg. life expectancy (years)")
g.set_title("Income and life expectancy in the US")
Text(0.5, 1.0, 'Income and life expectancy in the US')
Bonus: stack and unstack¶
A really clear overview here
gap_stack = gap_wide.stack("country")
gap_stack
# gap_stack.columns
| measure | gdpPercap | lifeExp | pop | |
|---|---|---|---|---|
| year | country | |||
| 1952 | Afghanistan | 779.445314 | 28.801 | 8425333.0 |
| Albania | 1601.056136 | 55.230 | 1282697.0 | |
| Algeria | 2449.008185 | 43.077 | 9279525.0 | |
| Angola | 3520.610273 | 30.015 | 4232095.0 | |
| Argentina | 5911.315053 | 62.485 | 17876956.0 | |
| ... | ... | ... | ... | ... |
| 2007 | Vietnam | 2441.576404 | 74.249 | 85262356.0 |
| West Bank and Gaza | 3025.349798 | 73.422 | 4018332.0 | |
| Yemen, Rep. | 2280.769906 | 62.698 | 22211743.0 | |
| Zambia | 1271.211593 | 42.384 | 11746035.0 | |
| Zimbabwe | 469.709298 | 43.487 | 12311143.0 |
1704 rows × 3 columns
# gap_stack['pop']
# gap_stack[gap_stack['year'] == 2007]
gap_unstack = gap_stack.unstack("year")
gap_unstack
| measure | gdpPercap | ... | pop | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| year | 1952 | 1957 | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | ... | 1962 | 1967 | 1972 | 1977 | 1982 | 1987 | 1992 | 1997 | 2002 | 2007 |
| country | |||||||||||||||||||||
| Afghanistan | 779.445314 | 820.853030 | 853.100710 | 836.197138 | 739.981106 | 786.113360 | 978.011439 | 852.395945 | 649.341395 | 635.341351 | ... | 10267083.0 | 11537966.0 | 13079460.0 | 14880372.0 | 12881816.0 | 13867957.0 | 16317921.0 | 22227415.0 | 25268405.0 | 31889923.0 |
| Albania | 1601.056136 | 1942.284244 | 2312.888958 | 2760.196931 | 3313.422188 | 3533.003910 | 3630.880722 | 3738.932735 | 2497.437901 | 3193.054604 | ... | 1728137.0 | 1984060.0 | 2263554.0 | 2509048.0 | 2780097.0 | 3075321.0 | 3326498.0 | 3428038.0 | 3508512.0 | 3600523.0 |
| Algeria | 2449.008185 | 3013.976023 | 2550.816880 | 3246.991771 | 4182.663766 | 4910.416756 | 5745.160213 | 5681.358539 | 5023.216647 | 4797.295051 | ... | 11000948.0 | 12760499.0 | 14760787.0 | 17152804.0 | 20033753.0 | 23254956.0 | 26298373.0 | 29072015.0 | 31287142.0 | 33333216.0 |
| Angola | 3520.610273 | 3827.940465 | 4269.276742 | 5522.776375 | 5473.288005 | 3008.647355 | 2756.953672 | 2430.208311 | 2627.845685 | 2277.140884 | ... | 4826015.0 | 5247469.0 | 5894858.0 | 6162675.0 | 7016384.0 | 7874230.0 | 8735988.0 | 9875024.0 | 10866106.0 | 12420476.0 |
| Argentina | 5911.315053 | 6856.856212 | 7133.166023 | 8052.953021 | 9443.038526 | 10079.026740 | 8997.897412 | 9139.671389 | 9308.418710 | 10967.281950 | ... | 21283783.0 | 22934225.0 | 24779799.0 | 26983828.0 | 29341374.0 | 31620918.0 | 33958947.0 | 36203463.0 | 38331121.0 | 40301927.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| Vietnam | 605.066492 | 676.285448 | 772.049160 | 637.123289 | 699.501644 | 713.537120 | 707.235786 | 820.799445 | 989.023149 | 1385.896769 | ... | 33796140.0 | 39463910.0 | 44655014.0 | 50533506.0 | 56142181.0 | 62826491.0 | 69940728.0 | 76048996.0 | 80908147.0 | 85262356.0 |
| West Bank and Gaza | 1515.592329 | 1827.067742 | 2198.956312 | 2649.715007 | 3133.409277 | 3682.831494 | 4336.032082 | 5107.197384 | 6017.654756 | 7110.667619 | ... | 1133134.0 | 1142636.0 | 1089572.0 | 1261091.0 | 1425876.0 | 1691210.0 | 2104779.0 | 2826046.0 | 3389578.0 | 4018332.0 |
| Yemen, Rep. | 781.717576 | 804.830455 | 825.623201 | 862.442146 | 1265.047031 | 1829.765177 | 1977.557010 | 1971.741538 | 1879.496673 | 2117.484526 | ... | 6120081.0 | 6740785.0 | 7407075.0 | 8403990.0 | 9657618.0 | 11219340.0 | 13367997.0 | 15826497.0 | 18701257.0 | 22211743.0 |
| Zambia | 1147.388831 | 1311.956766 | 1452.725766 | 1777.077318 | 1773.498265 | 1588.688299 | 1408.678565 | 1213.315116 | 1210.884633 | 1071.353818 | ... | 3421000.0 | 3900000.0 | 4506497.0 | 5216550.0 | 6100407.0 | 7272406.0 | 8381163.0 | 9417789.0 | 10595811.0 | 11746035.0 |
| Zimbabwe | 406.884115 | 518.764268 | 527.272182 | 569.795071 | 799.362176 | 685.587682 | 788.855041 | 706.157306 | 693.420786 | 792.449960 | ... | 4277736.0 | 4995432.0 | 5861135.0 | 6642107.0 | 7636524.0 | 9216418.0 | 10704340.0 | 11404948.0 | 11926563.0 | 12311143.0 |
142 rows × 36 columns
